Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 6

Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 1
... et al., 19 99) 10 0! Table 13 : Changes to ID2 protocol for expression and purification of ID2 and ID3 mutants 10 3! Table 14 : ID1 & ID2 constructs and their theoretical biochemical ... al., 19 90, Biggs, et al., 19 92, Christy, et al., 19 91, Riechmann, et al., 19 94, Sun, et al., 19 91) The mammalian family consisted of four members, namely ID1, ID2, ID3 and ID4...
Ngày tải lên: 09/09/2015, 10:20

Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 2
... cDNA (bp) AA start AA end #AA pI MW (kDa) 4 02 177 24 6 339 21 9 28 8 Construct 24 1 24 134 82 82 113 82 82 134 59 82 113 73 96 7.8 6.1 8.8 9 .2 6.1 8.8 14.9 6.8 9.3 12. 7 8.3 10.8 Full Length HLH24- 82 ... structure of ID2 3) Analyze the structure of ID2 and look at similarities and differences to other HLH-containing proteins including ID3 ! 20 ! 4) Determine differenc...
Ngày tải lên: 09/09/2015, 10:20

Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 3
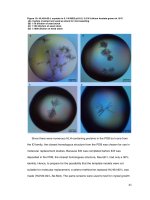
... marker (lane M, kDa) N-HLH82-L (gel A, lane 1), HLH24-82-L (gel B, lane 2), HLH24-82-L-Se-Met (gel C, lane 3) ! 38 ! 3. 3 Protein Identification The purified samples were excised from the gel and analyzed ... SILSLQASEF PSELMSNDSK ALCG Match to: Q53H99_HUMAN Score: 17248 Inhibitor of DNA binding variant (Fragment).- Homo sapiens (Human) Found in search of C:\Documents and Set...
Ngày tải lên: 09/09/2015, 10:20
Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 4
Ngày tải lên: 09/09/2015, 10:20

Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 6
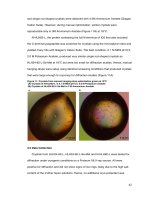
... solution and was abandoned in favour of the higher resolution dataset of the longer form of ID2, N-HLH82-L The strategy for running MR was to use 1-2 ensembles with the Se-Met dimer and monomer ... was composed of a 4-helix bundle refined to 2.1Å with an Rfree value of 25% and no Ramachandran outlier (Table 10, Figure 13) PyMol (DeLano, 2002) was used for generating all th...
Ngày tải lên: 09/09/2015, 10:20
Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 5
Ngày tải lên: 09/09/2015, 10:20
Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 7
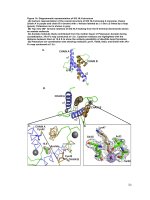
... mutagenesis was done on Y71 and Q76 Single mutations to change the size and charge of the residues were Y71A, Y71F, Q76A and Q76D (Table 5) Since these interactions were not found in any of the non-ID homodimers ... consisting of residues M39, L46, L49, M62, I69, I72, L75 (Figure 16A), all of which had equivalent buried residues in the structures of E 47 (Ellenberger, et al.,...
Ngày tải lên: 09/09/2015, 10:20
Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 8
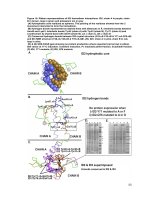
... red arrow) and ID3-Q66K did not produce any soluble protein, thus suggesting the importance of Q66 specifically to ID3’s stability and solubility Figure 17: Loop region mutants of ID2 and ID3 SDS-PAGE: ... ID2 and ID3 and its mutation in ID1 led to a complete loss of binding to MYOD1 and E47 (Pesce, et al., 1993) Since prolines were known to disrupt secondary structures, s...
Ngày tải lên: 09/09/2015, 10:20

Structural characterization and biochemical analysis of ID2, an inhibitor of DNA binding 9
... Edlund, T and Walker, M D ( 199 1) "Distribution and characterization of helix-loop-helix enhancer -binding proteins from pancreatic beta cells and lymphocytes", Nucleic Acids Res, 19, 3 893 -9 Atchley, ... Hernandez, M C., Kuo, W L and Israel, M A ( 199 8) "The mouse Id2 and Id4 genes: structural organization and chromosomal localization", Gene, 222, 2 29- 35 80 Martinsen...
Ngày tải lên: 09/09/2015, 10:20

Tài liệu Báo cáo khoa học: Cloning, characterization and expression analysis of interleukin-10 from the common carp, Cyprinus carpio L. docx
... to their positions The arrowheads depict the residues important for the structural core of the IL-10 gene The underlined amino acid residues are the signal sequences of the respective genes The ... conclusion, the IL-10 gene from carp has been isolated and its genomic structure and expression analysis investigated This work will pave the way for further inves...
Ngày tải lên: 20/02/2014, 02:21

Báo cáo khoa học: Proteomic and biochemical analysis of 14-3-3-binding proteins during C2-ceramide-induced apoptosis pot
... comprehensive proteomics analysis of 14-3-3-binding proteins under physiological conditions as compared with apoptosis stimulation, with the aim of increasing our knowledge of the role of 14-3-3 proteins ... changes in 14-3-3-binding patterns in HeLa cells during C2-ceramideinduced apoptosis With the aim of further analyzing the role of 14-3-3 proteins in apopto...
Ngày tải lên: 06/03/2014, 22:21

Báo cáo khoa học: Isolation, characterization and expression analysis of a hypoxia-responsive glucose transporter gene from the grass carp, Ctenopharyngodon idellus potx
... 5¢-CCTGATCGACGCACGAGT-3¢ and GT1-R, 5¢-TTTTGCAAGTCATAGTAATCAGTTT-3¢ for GTcDNA1 (2150 bp); and GT2-F, 5¢-CACCAGCAACTAC CTGATCGA-3¢ and GT2-R, 5¢-CACAAAATATGCTT CCAAGTGC-3¢ for GT-cDNA2 (3043 bp) RNA isolation and ... revealed two putative polyadenylation (ATTAAA) signals: one is located 18 bp upstream from the poly (A) of GT-cDNA1 and another is located 11 bp upstream from...
Ngày tải lên: 17/03/2014, 03:20

Báo cáo khoa học: Characterization and expression analysis of the aspartic protease gene family of Cynara cardunculus L. docx
... pattern of expression of a gene [34–39] To evaluate the relevance of the leader intron in cardosin expression, we deleted it from the 5¢-flanking region of the genes (Fig 8) The deletion of the cardosin ... region of the cardosin B gene (from ) 147 bp to + 238 bp) .The 529 bp of the promoter region of the cardosin A gene that is relevant for gene...
Ngày tải lên: 30/03/2014, 09:20

Báo cáo khoa học: Dual expression of mouse and rat VRL-1 in the dorsal root ganglion derived cell line F-11 and biochemical analysis of VRL-1 after heterologous expression pptx
... that the F-11 cells not only express the rat VRL-1, but also the mouse VRL-1 and that the mouse variant is derived from the F-11 parental cell line N18TG2 This finding is of interest when VRL-1 ... A, B and D of F-11 cells again separated into two bands The slower migrating band always corresponded to the rat brain control, and the faster migr...
Ngày tải lên: 31/03/2014, 07:20

Báo cáo y học: " Complete coding sequence characterization and comparative analysis of the putati" docx
... drafted the manuscript SP and KS participated in the sequence alignment PL and YP participated in the design of the study and performed the data statistical analysis YP conceived of the study in ... only 64% sequence identity with the other HRV-Cs HRV-CU072 coding sequence analysis To investigate the molecular characteristics of the putative new HRV-C...
Ngày tải lên: 11/08/2014, 21:21